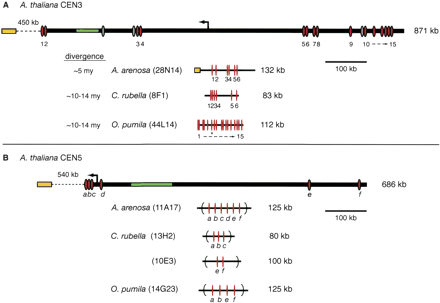
Multispecies comparisons of regions homologous to A. thaliana pericentromeres. Scale drawings of (A) A. thaliana peri-CEN3 (chromosome III bases 14,096,340–14,967,565), and sequenced BACs from A. arenosa, C. rubella, and O. pumila. (B) A. thaliana peri-CEN5 (chromosome V bases 12,566,510–13,252,797), and homologous BACs identified by hybridization, from A. arenosa, C. rubella, and O. pumila (complete list, Supplemental Table 2). Estimated times of divergence (millions of years, Myr) are shown (Koch et al. 2001). Dotted lines, estimated distance (not to scale) to the centromere satellite array (yellow boxes); (solid arrows) centromere boundary as defined by recombination (Copenhaver and Preuss 1999); (green boxes) 5S rDNA array; (red bars and ovals) expressed A. thaliana genes present in multiple species; (gray ovals) intact A. thaliana-specific genes; (left to right) At3g42050, At3g42150, and At3g42786). (*) Gene homologous to A. thaliana chromosome I expressed gene (At1g63270); (dashed arrows) all genes in the interval are present; (parentheses in B) undetermined gene order.











