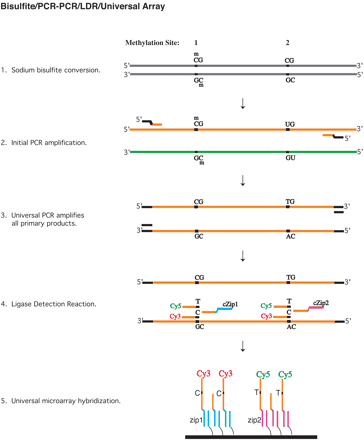
Schematic diagram of the assay. Two hypothetical CpG dinucleotide sites 1 and 2 are designated as methylated and unmethylated, respectively. Sodium bisulfite converts unmethylated, but not methylated, cytosines into uracils. This conversion renders the genomic DNAs into two asymmetrical, noncomplementary strands, and only one designated bisulfite-modified strand (highlighted in orange) is amplified and analyzed. In the initial amplification, PCR primers are designed with a gene-specific 3′ portion and an upstream universal sequence (highlighted in black). This universal sequence is used as a PCR primer in the subsequent PCR to simultaneously amplify all the primary amplicons (for ease of illustration, only one amplicon is shown). LDR is performed in a multiplex fashion with three primers (two discriminating and one common primers) interrogating each of the selected CpG sites. The discriminating primers contain a 5′ fluorescent label and a 3′ discriminating nucleotide to determine either methylated (with 5′ Cy3 and 3′ C) or unmethylated (with 5′ Cy5 and 3′ T) cytosines. The common primers bear a 5′ phosphate and a 3′ unique zip-code complement sequence (e.g., cZip1 and cZip2). Ligation occurs only if the nucleotides at the ligation junction are perfectly base-paired with a complementary template and the ligation products are captured onto a Universal microarray with prespotted zip-codes (addresses). For example, address Zip1 identifies methylated cytosine in methylation site 1, and address Zip2 identifies unmethylated cytosine in methylation site 2. Three Universal microarray addresses are assigned for each promoter region, and each address is double-spotted to ensure the quality of array fabrication and oligonucleotide hybridization efficiency.











