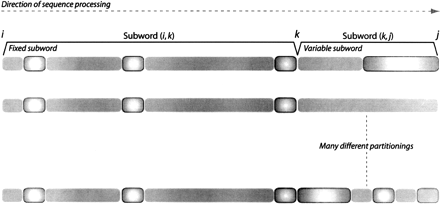
Many different partitionings share common subparts. To compute any partitioning with a split at k, the best partitioning for (i, k) and for (k, j) must be known. Since there are many ways to partition the sequence with a split at k, we only need to recursively evaluate a subpartitioning for subword (i, j) and (k, j) once. In all cases where we need the optimal solution for these subwords again, we refer to the pre-computed result instead of considering all further possible partitionings of that subword.











