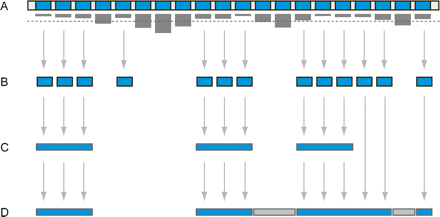
The problem of sequence similarity in tiling genomic DNA. (A) The level of similarity of oligonucleotide sequences to the remainder of the genome is represented by descending bars, where longer bars indicate more redundant sequences. If the redundancy exceeds a given threshold, indicated by the dashed line, the sequence is omitted from the tile path (B). Avoiding redundant or repetitive sequences inhibits adequate tiling of the sequence (C). Here, the level of non-repetitive sequence coverage decreases as the minimum tile size increases. At this point it also becomes necessary to use approximations that identify instances of known DNA transposons, retroelements, satellites, and other repetitive sequences, rather than calculating an explicit measure of sequence similarity. (D) In order to recover a higher percentage of non-repetitive DNA, tiling algorithms can be devised that incorporate some redundant sequences (gray) in an optimal fashion, which balances the cost of inclusion against the gain in sequence coverage.











