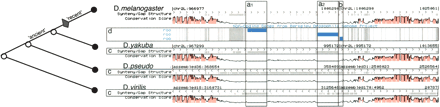
An alignment of four Drosophila sequences shown in the K-browser. The species are labeled by D. melanogaster, D. yakuba, D. pseudo (pseudoobscura), D. virilis. Each genome track has a conservation score track (pink) and (c) a gap track (gaps are demarcated in gray). The gaps in three genomes support the correct boundaries of the D. melanogaster insertion regions (a1, a2, and b). The insertion regions match the TE annotations (blue) in the BDGP noncoding gene track (d). The tree on the left-hand side depicts the phylogeny relationships between the species. The diamond shows the branch on which the transposon replications (a1) and (a2) occurred. The “ancient” branch is that on which replication (b) occurred, as indicated by the gaps in D. pseudoobscura and D. virilis but not in D. yakuba.











