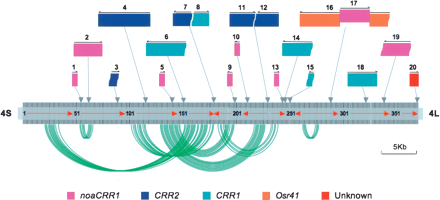
Arrangement and rearrangement of centromeric repeats in the core region of rice centromere 4. The intact LTR-retrotransposons and solo LTRs are represented by color-shadowed boxes, whereas the truncated retrotransposons are represented by color-shadowed boxes with one or both curved ends. The five families/subfamilies of retrotransposons are marked by five different colors. (Arrows and numbers above the shadowed boxes) Orientations and order of LTR-retrotransposons in the region analyzed, (dark vertical lines) CentO satellite monomers, (vertical arrows connected to the boxes) positions of LTR-retrotransposons, (red arrows) orientation of CentO blocks separated by LTR-retrotransposons. The most related pairs of monomers distributed in corresponding duplicated segments in conserved order (in the same or opposite orientations) are connected by curved green lines.











