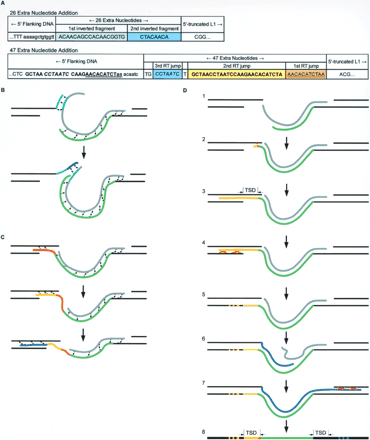
(A) 5′ junctions of insertions containing 26 and 47 extra 5′ nucleotides. Their likely formation is depicted in B and C, respectively. TSD bases are in lower case. (Flanking host DNA) Solid black lines; (L1 RNA) gray line; (nascent cDNA strand) light-green line; (conceptual base pairing) dotted black lines. (B) A schematic of the “dual inversion” mechanism that created the 26 extra nucleotides between the flanking host DNA and the 238-bp 5′-truncated insert. Extra nucleotides are a product of two successive twin priming reactions (in A, highlighted in light and dark blue). Likely, the first twin priming reaction was initiated using a 1-bp MH, synthesizing the first inverted fragment of 18 bp (light blue). Then, the RT underwent a template jump, creating the second inverted fragment of 8 bp (dark blue). (C) Successive L1 RT template jumps created an insertion of 47 nt in a 368-bp de novo integrant. The 47-nt addition contains three overlapping regions of homology to the host DNA immediately 5′ to the insertion site (in A, underlined orange, bold and yellow, and italicized and blue). When L1 RT reached the end of the RNA template at the end of a TPRT reaction, it likely template jumped from L1 RNA to the host DNA strand. Nascent cDNA was elongated by 11 bases (orange) copying a portion of host DNA. A second RT jump occurred, copying the same region of host DNA for 26 nt (yellow). One base was added (data not shown), and a third RT template jump occurred extending for seven more nucleotides (blue). Two bases were added (data not shown), and the structure was resolved, creating an addition of 47 nt. (D) Model for L1 integration. (Flanking host DNA) Solid black lines; (L1 RNA) gray line; (nascent cDNA strand) green line; (nascent 2nd strand) blue line; (homologous recombinational repair) orange crossed lines; (recombination products [no sequence changes expected]) stippled black/yellow and black/blue lines; (5′MH-guided base pairing) dotted red line; (TSD) target site duplication. See Discussion for detailed explanation.











