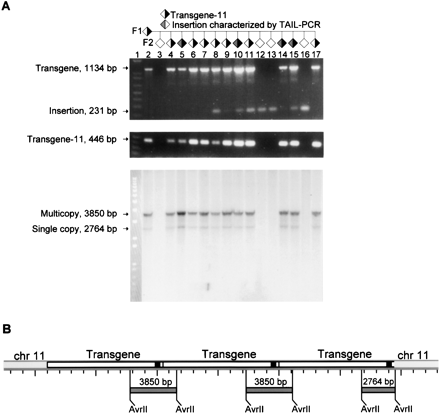
Characterization of a pedigree segregating transgene-11. (A)F1 mouse and its 15 offspring (top pedigree schematic) were genotyped by PCR with L16045(30) and mlessfor2 oligos (top gel) to reveal 11/15 F2s carrying transgene (1134 bp) and 7/15 carrying de novo inserts (231 bp). Minus DNA control was negative; de novo insertion presence was confirmed with different oligos and independently isolated tail DNA stocks (data not shown). All transgene inheritance was accounted for by transgene-11-specific PCR (middle gel, 446 bp). Southern blot (bottom gel) of 10 μg of tail DNA, digested with AvrII, and probed with a 32P-labeled 376-bp probe showed multicopy (3850 bp) and single-copy (2764 bp) fragments expected for AvrII digest of transgene-11, shown in B. No other fragments were detected, suggesting that the frequency of de novo insertions is less than one copy/cell. Four F2s contained inserts characterized by TAIL–PCR (gray shading in pedigree schematic). A 1-kb Plus DNA Ladder is shown in lane 1 of all gels. (B) A hypothetical multicopy transgene-11 array with relevant AvrII sites and the 376 bp probe (black rectangle).











