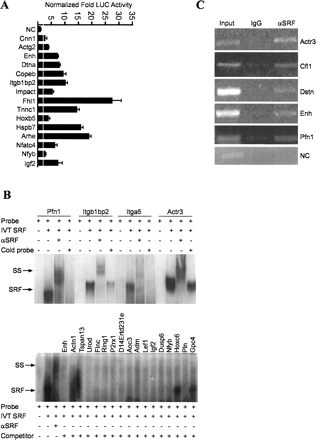
Functional validation studies of CArG-containing sequences. (A) Representative luciferase assay results for a sample of computer-predicted CArG sequences (13 novel and two known) in C2C12 myoblasts. The white vertical line across bars indicates the experimentally defined threshold for scoring a target CArG sequence as positive over the value obtained from a collection of negative controls (see Methods); (NC) negative control is the tk promoter-linked luciferase plasmid only. (B) Representative in vitro SRF-binding assays for predicted CArG sequences. (Top panel) The results of radiolabeled target sequences binding to in vitro translated (IVT) SRF. Note supershift of each nucleoprotein complex with antibody to SRF. Addition of unlabeled target DNA attenuates the nucleoprotein signal. (Bottom panel) A cold competition EMSA in which a radiolabeled probe containing the CArG sequence CCTTATTTGG was incubated with IVT SRF in the absence or presence of a molar excess of each target CArG-containing sequence. The results indicate that all target sequences except Hoxc6 and Gpc4 compete with labeled CArG probe for binding to IVT SRF, thus reducing the signal intensity of the nucleoprotein complex. The smearing below Actn1 and Tspan13 is an artifact of the gel. (C) ChIP assay results for a select group of novel SRF targets showing an enriched PCR product from cross-linked DNA immunoprecipitated with SRF antibody. No detectable PCR product is seen for a region of a negative control sequence (NC) corresponding to the Myocd gene, which does not contain any CArG sequences. Moreover, little or no amplified product is observed for any of the CArG targets when an IgG control antibody is used to immunoprecipitate cross-linked DNA.











