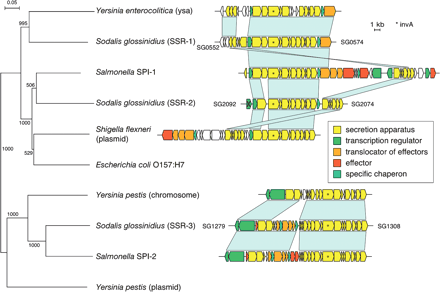
Phylogenetic relationship shared between Sodalis' three TTSS structures and those characterized from pathogenic microbes. The tree was based on amino acid sequence alignments of the invA gene product. Scale bars representing branch lengths and bootstrap values are displayed at each internal node. Genes associated with each cluster are depicted as arrows that indicate the direction of transcription. The arrows with a cross denote pseudogenes. The light blue bars between loci indicate the regions of sequence similarity and gene order conservation. The different colors depict the functional roles of their putative products.











