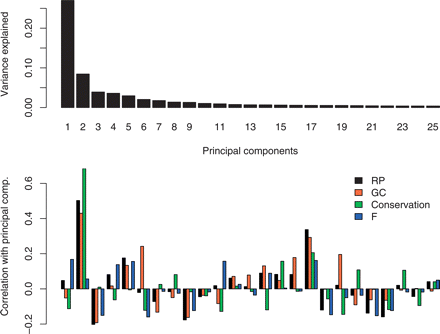
Figure 3.
Share of variance explained by each of the first 25 principal components of the RP training data word frequencies (top) and correlation of RP score, GC content, conservation, and the residuals F with each principal component (bottom).











