Table 3.
Promoters bound by both E2F1 and AP-2α
Click on table to view larger version.
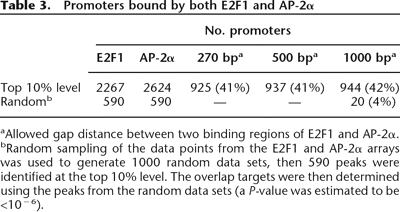
aAllowed gap distance between two binding regions of E2F1 and AP-2α.
bRandom sampling of the data points from the E2F1 and AP-2α arrays was used to generate 1000 random data sets, then 590 peaks were identified at the top 10% level. The overlap targets were then determined using the peaks from the random data sets (a P-value was estimated to be <10−6).











