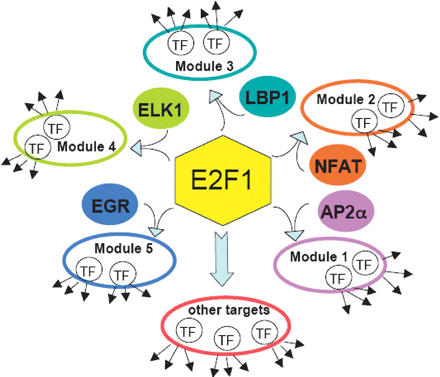
E2F1 cooperates with and regulates other transcription factors. Shown is a schematic indicating the five different modules identified in this study using E2F1 ChIP–chip data. DAVID analysis of the OMPGProm database-identified promoters (Supplemental Table S3) revealed that transcription factors are a predominant category of target genes in all five E2F1 ChIPModules: Nucleic acid binding proteins constituted ∼20% of ChIPModule 1, 26% of ChIPModule 2, 10% of ChIPModule 3, 26% of ChIPModule 4, and 22% ChIPModule 5.











