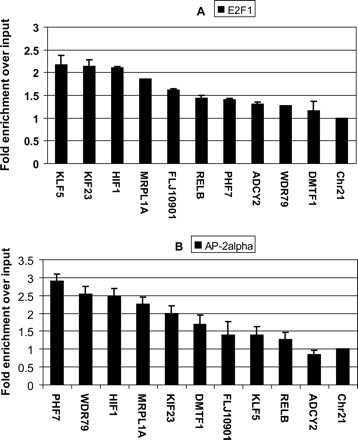
PCR confirmations of E2F1 (A) and AP-2α (B) binding to a set of 10 randomly selected promoters predicted by our ChIPModules approach from the OMGProm data set. A region of chromosome 21 is used as a negative control. All fold enrichments were compared to the enrichment of the total input and normalized to the negative control. The signals were within the linear range of the assay, providing a semiquantitative analysis. The error bars were calculated based on the standard deviation from two samples of confirmation.











