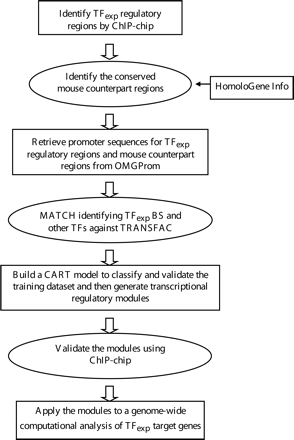
Flow chart showing the ChIPModules approach. Shown is a schematic indicating the steps needed to develop a database of target promoters for a particular site-specific human transcription factor. The approach begins with a set of experimentally defined binding sites (TFexp), refines the set to include only those sites conserved in the orthologous mouse promoters, searches for nearby binding sites for other factors, builds a CART model to generate a high confidence set of co-occurring binding sites, validates the colocalization of the factors using additional ChIP–chip assays, and then searches for the validated ChIPModules in a large promoter database. A detailed description of each step can be found in Supplemental Figure S5.











