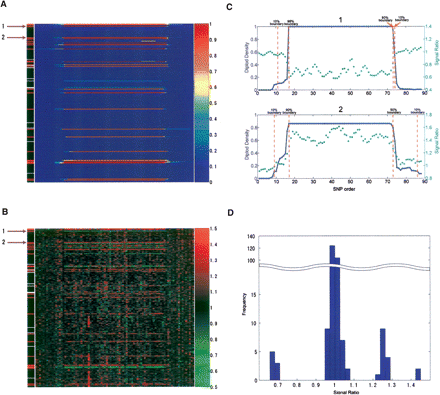
CNV boundary determination. In A,B,C, the x-axis is the sequential order of the SNPs both within and outside the CNV region; the y-axis represents individual samples. In D, the x-axis represents the intensity ratio, and the y-axis is the sample frequency. (A) Diploid density distribution for HapMap CNVID 1166 at chr22:23932716–24371067. In the left-most column, CNV samples detected by the algorithm are shown in red, the diploid samples selected by the maximum clique algorithm are shown in black, and samples that display the intensity trend but do not meet the CNV extraction criteria are shown in white. (B) Median ratio distribution in the same region as shown in A. The median ratios were calculated based on the diploid samples with the same genotype. The similar pattern with A indicates that the CNV regions were successfully detected by the algorithm. (C) The diploid density (blue solid line) and median ratio (green dotted line) smoothed with a 10 probe window of sample 1 (top graph) and sample 2 (bottom graph) indicated by the purple arrows in A and B. The 10% and 90% boundaries are shown as dashed red lines. (D) The intensity ratio histogram of all 270 samples in the same CNV region depicted in A, B, and C shows clear clusters that correspond to one, two, three, and four copies of the region. The histogram is compressed in the middle range of the y-axis as represented by the wavy double line.











