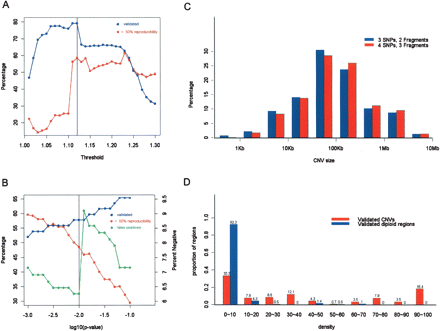
Parameter tuning. Several parameters were optimized for SW-ARRAY and CNV extraction including (A) intensity ratio threshold, (B) statistical significance, (C) number of SNPs and restriction fragments required for calling a CNV, and (D) density cutoff (the fraction of positive pairwise comparisons necessary for calling a CNV). For each parameter, CNVs were called from pairwise comparisons between NA15510 and NA10851 (A,B,C) or population-wide comparisons (D) for each sample. In A and B, the percentage of CNVs called more than half of the time (red line) is compared to the percentage that have been positively validated (blue line), or negatively validated, false positives (green line in B with the y-axis on the right-hand side). The final values chosen were 1.12 for the intensity ratio threshold, and 0.01 for the P-value (indicated by the vertical black lines). In C, the size distribution of CNVs detected using the 3 SNPs:2 fragments criterion (with a mean length of 300 kb) is compared to the 4 SNPs:3 fragments criterion (mean length 326 kb). In D, the number above each bar is the percentage of validated CNVs and validated diploid regions within certain density bins. A 10% density cutoff was chosen for further analyses. For any given cutoff, a false positive is defined as the percentage of all validated diploid regions that are incorrectly called as a CNV with a density that is greater than the cutoff; false negative is defined as the percentage of validated CNVs that are missed because their density is lower than the cutoff.











