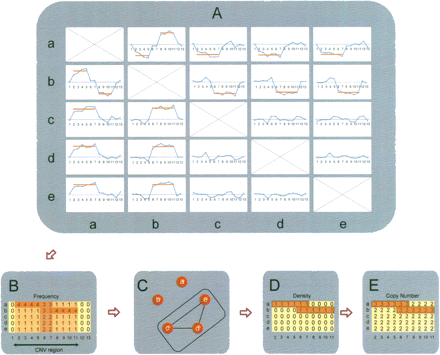
Overview of copy number inference. (A) Pairwise comparisons of five different DNA samples (a–e) in a given candidate CNV region. The x-axis represents the SNP positions, and the blue lines are log2 signal intensity ratios for any given pair. The red line indicates the significant CNVs detected by SW-ARRAY. (B) Summary of the comparisons of any given sample to the remaining four samples. Based on the physical location of copy number changes, the frequencies are calculated for each sample, and consecutive CNV regions are extracted. Each row represents a single sample, and each column represents the frequency of a given SNP. The frequency of a particular SNP is the number of times that it is called a CNV in all four pairwise comparisons. (C) Graph theory (the maximum clique algorithm) is applied to the frequency summarization results presented in B. In this example, samples c, d, and e, which have the lowest frequency and represent the maximum clique, are defined as the diploid group. (D) Density (the proportion of comparisons where a CNV is called) is calculated based on the diploid samples found by the maximum clique algorithm, and the boundary of the CNV region in each nondiploid sample is determined. (E) Copy number is determined based on the median ratio of each CNV region.











