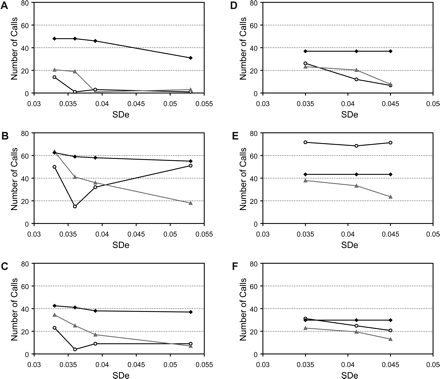
Comparison of CNVfinder with SW-ARRAY. CNV calling using CNVfinder (A), SW-ARRAY-permissive (B), and SW-ARRAY-stringent (C) in five replicate experiments. The number of regions called in three or four replicates (black diamonds), one or two replicates (gray triangles), or in none (white circles) of the other replicates is plotted against global SDe values. (D,E,F) CNV calling using CNVfinder (D), SW-ARRAY-permissive (E), and SW-ARRAY-stringent (F) in triplicate experiments of 10 different cell line DNAs. The replicate experiments were pooled into three different sets X, Y, and Z (see Table 3). The number of regions called in the two other replicates (black diamonds), one other replicate (gray triangles), or in none (white circles) is then plotted against the average of the global SDe values for each set.











