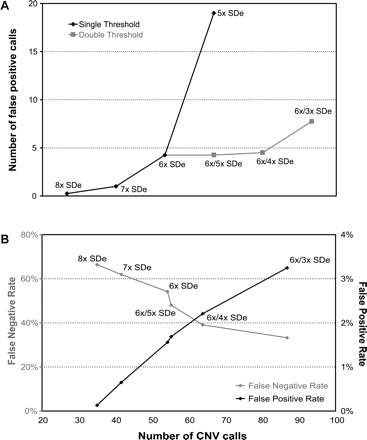
CNVfinder performance using varying SDe multiplier thresholds in WGTP array validation experiments. (A) Number of false-positive calls in self–self experiments in relation to SDe multiplier thresholds. Various single/dual SDe multiplier thresholds were applied to four replicate self–self experiments and the mean number of calls calculated. (Black diamonds) mean number of regions called by single SDe multiplier thresholds, (gray squares) mean number of regions called by dual SDe multiplier thresholds. (B) False-positive and false-negative rates against the number of CNVs called for varying single/dual SDe multiplier thresholds in NA15510 versus NA10851 experiments. False-negative and false-positive rates are based on the quantitative PCR results from 154 sampled clones.











