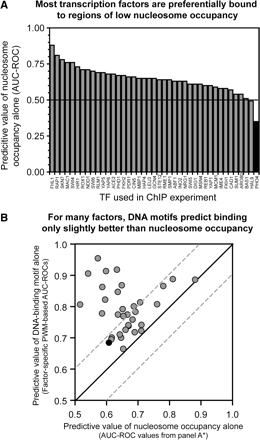
For many sequence-specific DNA-binding proteins, nucleosome occupancy alone predicts promoters bound in vivo almost as accurately as DNA sequence. (A) Most transcription factors are preferentially bound to regions of relatively low nucleosome occupancy. ROCs were used to quantitate the value of low nucleosome occupancy in predicting the in vivo distribution of the indicated transcription factors. ChIP-enriched sequences (Lee et al. 2002) were defined at 10% FDR. Only the 41 ChIPs yielding at least 20 enriched sequences were analyzed. Pho4 (black) appears to be significantly associated with regions of higher nucleosome occupancy, possibly because of different growth conditions under which ChIP-chip and nucleosome data were collected. (B) Motifs derived from bound sequences often predict binding only slightly better (within an AUC-ROC of 0.1) than does nucleosome occupancy alone. Of the 41 factors in A, a significant DNA-sequence motif could be derived for the 34 plotted here (see Supplemental Table 2 for tabular data). PWMs were used to calculate occupancy scores for every intergenic region. ROCs were then used to quantitate the ability of the occupancy scores derived from the factor-specific motifs to predict the in vivo distribution of the corresponding factor (y-axis). On the x-axis are the nucleosome-occupancy-based AUC-ROCs. AUC-ROC values under 0.5 were converted to (1 − AUC-ROC). Of the transcription factors, 17/34 fall between dashed lines, which indicate AUC-ROC values within 0.1. (Filled black circle) Pho4.











