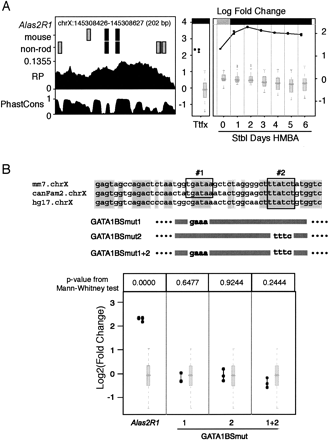
Features, validation, and mutagenesis of a preCRM in the Alas2 locus. (A) Features and activities of preCRM Alas2R1. The graph on the left shows important features of the preCRM: the chromosomal position and size, segments that match the consensus GATA-1 binding site (black rectangles) and matches to the weight matrix for the GATA-1 binding site that are not matches to the consensus (gray rectangles) in mouse and in non-rodent (non-rod) sequences, regulatory potential scores (RP), and phastCons scores (PhastCons) across the genomic interval. The two graphs on the right summarize the activities of the preCRMs (connected dark filled circles), represented as log2 fold change and compared with that of the preNeutrals (light gray box-plot). The graph labeled “Ttfx” plots the values of the transient transfection experiments in K562 cells. The seven columns in the rightmost graph show the results for the induction time course (Stbl Days HMBA) of pools of cells after site-directed integration of the test expression cassette in MEL_RL5 cells. Significant differences of preCRM activity compared with the distribution of activity measurements for preNeutrals are denoted by a black bar along the top of the graphs; otherwise the bar is gray. (B) Mutagenesis and tests of GATA motifs in Alas2R1. The alignment of a subregion of mouse Alas2R1 with dog (canFam2) and human (hg17) shows two conserved consensus GATA-1 binding sites (outlined). Alas2R1 was mutated in individual (BSmut1 and BSmut2) and both (BSmut1 + 2) GATA-1 binding sites, with the block substitutions shown beneath the alignment. The activity levels of expression plasmids carrying the wild-type and mutated Alas2R1 preCRM are shown in the bottom panel, using the same conventions as in A, except that the numerical P-values for a difference of the activity of the test construct compared with the activities of preNeutrals is given at the top of each column.











