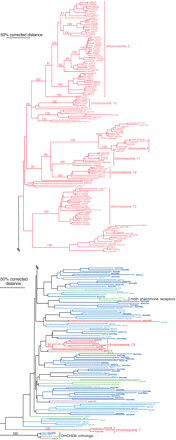
Phylogenetic relationships of the 170 AmOrs and fly and moth Ors. This corrected distance tree was rooted by declaring the DmOr83b protein and its orthologs in the other species (AgOr7, HvOr2, and AmOr2) as the outgroup, based on the position of DmOr83 at the base of the Or family in phylogenetic analyses of the entire superfamily in Drosophila (Robertson et al. 2003). Relatively closely related fly and moth Ors were removed to facilitate phylogenetic analysis and presentation, hence many of the lineage-specific subfamily expansions in these insects are not obvious (see Hill et al. 2002). Clusters of tandem arrays of AmOrs on particular chromosomes are indicated by vertical lines on the right. Numbers above branches are the percentage of 1000 neighbor-joining bootstrap replicates containing that branch and are shown only for major lineages including bee Ors. Symbols after the AmOr numbers: F indicates the missing genomic sequence was manually assembled to complete the gene model; N, the N terminus of the gene model is missing in a gap or is unidentifiable; C, the C terminus of the gene model is missing in a gap or is unidentifiable; and P, pseudogene with one or more in-frame stop codons, frame-shifting indels, or unacceptable intron splice sites.











