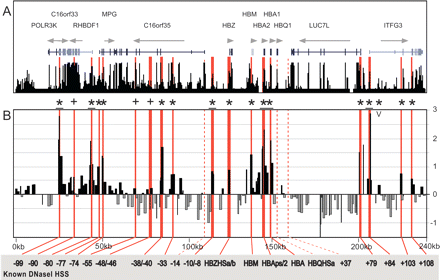
DNaseI hypersensitive site profile across the human α-globin locus. (A) Organization of the 12 genes across 240 kb, with wide and narrow bars representing translated and untranslated exons, respectively. The black bars represent the human/chimp/mouse/rat/dog/chicken overall conservation score quantified using phastCons (Siepel et al. 2005), derived using the MultiZ species alignment program (http://genome.ucsc.edu) (Hinrichs et al. 2006). (B) DNaseI sensitivity plot from K562 cell lines. Fold enrichment over non-enriched input is plotted (log2) against genomic position in kilobases. The plot represents the mean values of seven separate array experiments, each hybridized in duplicate as detailed in Methods. The width of each bar represents the width of each spotted PCR product on the array, with overlapping bars representing overlapping PCR products. Red bars represent known regulatory elements, while dashed red bars are known elements not represented on the array. (0k) Human chromosome 16 coordinate 17,913 (NCBI build 35). Significant enrichments (P ≤ 0.05) are indicated with an asterisk or asterisk + bar (at least two adjacent tiles), while crosses represent the three known hypersensitive sites that were enriched above baseline, but did not meet statistical criteria for significant enrichment. The “V” indicates the two novel significant sites identified within the region of the locus previously mapped by conventional techniques (Yagi et al. 1986; Higgs et al. 1990; Vyas et al. 1992; Hughes et al. 2005; D. Higgs, pers. comm.). Functional elements refer to kilobases relative to HBZ exon 1.











