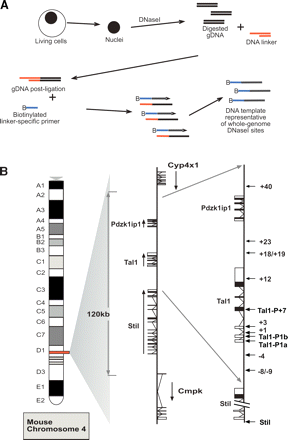
Protocol outline and map of the mouse Tal1 locus. (A) Diagrammatic representation of the protocol followed to obtain DNA template representative of whole-genome DNaseI hypersensitive sites. Nuclei are isolated and treated with DNaseI. DNA is then extracted and repaired to blunt ends with T4 DNA polymerase; (gDNA) digested, repaired genomic DNA. Following ligation to an asymmetric double-stranded linker, biotinylated primer extension is used followed by extraction of biotinylated primer-extended DNA using streptavidin beads. This template is then labeled and hybridized to genomic arrays as described in Methods. (B) Diagrammatic map of mouse chromosome 4 showing the organization of the three genes: Stil (Scl/Tal1 Interrupting Locus), Tal1, and Pdzk1ip1. In the middle panel, the direction of transcription is indicated with black arrows, while the locations of known Stil and Tal1 regulatory elements are indicated with arrows in the far right panel.











