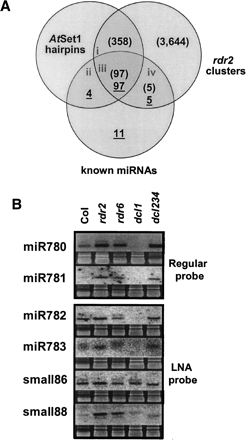
Estimating the minimal Arabidopsis miRNA population. (A) A Venn diagram to identify the number of miRNAs that may exist in Arabidopsis. Numbers in each section indicate sequences that satisfy each criterion, include AtSet1 hairpin folds (Jones-Rhoades and Bartel 2004), known miRNAs (underlined numbers), and genomic clusters of rdr2 MPSS signatures (numbers in brackets). AtSet1 and rdr2 clusters were considered matching if the start or end of either overlapped. Lowercase Roman numerals indicate sections from which sequences were selected in B. (B) Gel blots to evaluate the fraction of bona fide miRNAs in selected sectors. Small RNAs identified by rdr2 signatures in section (i) in A were randomly selected and evaluated by RNA gel blots to determine their presence/absence in the dcl1–7, dcl2/3/4, and rdr6 mutants. LNA probes were used for miR782, miR783, small86, and small88 (see Methods).











