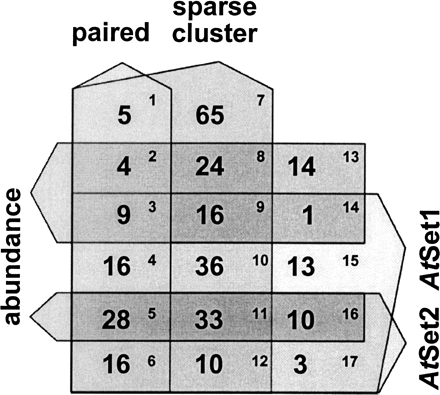
Use of rdr2 sequences to select miRNA candidates from previously identified wild-type small RNAs. Five-way Venn diagram of selection criteria for miRNAs. The number of distinct rdr2 MPSS signatures matching the criteria is indicated in each box numbered in upper right; only rdr2 signatures also found in the wild-type library are represented. The figure excludes 13,153 distinct signatures that did not pass any of the criteria (of which 1583 were found in both rdr2 and wild-type inflorescence libraries) and 54 matching to the criteria in the Venn which were present in rdr2 but not wild-type inflorescence (these 54 are included in Supplemental Fig. S2). The paired, sparse, abundance filters, and AtSet1 and AtSet2 filters are described elsewhere (Jones-Rhoades and Bartel 2004; Lu et al. 2005) but represent potential hairpin structures typical of miRNA precursors and conservation of those structures in rice, respectively.











