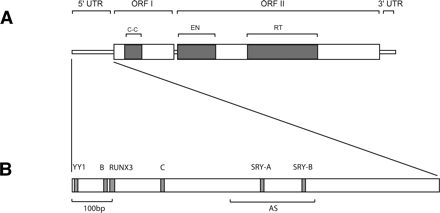
(A) Structure of a modern human full-length element. A full-length element is 6 Kb long and contains a 5′ untranslated region (5′UTR), two open-reading frames (ORFI and ORFII), and a 3′UTR. The 5′UTR has a regulatory function (Swergold 1990; Minakami et al. 1992). ORFI encodes a protein with nucleic acid-binding properties that can also act as a nucleic acid chaperone (Martin et al. 2000; Martin and Bushman 2001). ORFIp also contains a coiled-coil domain (C-C) that mediates interaction of ORFIp with itself (Martin et al. 2000). ORFII encodes a protein with endonuclease (EN) (Feng et al. 1996) and reverse transcriptase (RT) activity (Mathias et al. 1991). The 3′UTR contains a conserved poly-purine tract (Howell and Usdin 1997). Genomic copies of L1 are typically flanked by an A-rich tail at their 3′ end. (B) Functional motifs in the 5′UTR of a modern L1 element (L1PA1). The first 100 bp (100bp) of the 5′UTR was shown to be critical for transcription (Swergold 1990). The 5′UTR contains a YY1 binding site that plays an important role in transcription initiation (Athanikar et al. 2004), a functional RUNX3 binding site (Yang et al. 2003), two functional SRY-related transcription factor binding sites (SRY-A and SRY-B) (Tchenio et al. 2000), and two cellular factor-binding motifs (B and C) (Minakami et al. 1992). The 5′UTR also contains an antisense promoter (AS) between positions 400 and 600 that can drive transcription of adjacent cellular genes (Speek 2001).











