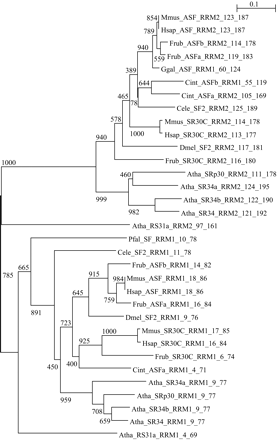
Evolutionary relationship among the RNA-recognition motifs (RRM) of members of the family SRp30c-ASF for several eukaryotes, i.e., human (Hsap), mouse (Mmus), chicken (Ggal), Fugu (Frub), Ciona (Cint), fruit fly (Dmel), C. elegans (Cele), Arabidopsis (Atha), and Plasmodium (Pfal) (for simplicity only one rodent, one teleost, and one insect are shown). Amino-acid positions of each domain within the protein are also indicated in the domain identification. The unrooted Neighbor-Joining phylogenetic tree was generated using ClustalW (1000 bootstraps) based on amino-acid alignment generated by T-Coffee. Bootstrap values are shown. Branch lengths are scaled in arbitrary units. RRM1 in Ggal_ASF and Cint_ASFb corresponds to RRM2 in the other proteins as their sequences are truncated in the N-terminal. Pfal_SF is found to have only one RRM. Atha_RS31A can be technically considered an ortholog of the Hsap_SR30C (reciprocal BLAST hit) but exhibits a considerably lower degree of identity (36%) with the human factor than its Arabidopsis paralogs (e.g., 53% for Atha_SRp30).











