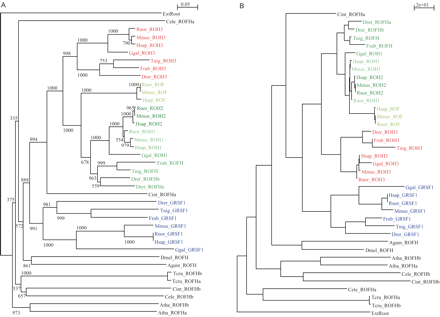
Evolutionary relationship among the protein members of hnRNP F/H family in several eukaryotes, i.e., human (Hsap), mouse (Mmus), rat (Rnor), chicken (Ggal), Fugu (Frub), zebrafish (Drer), Tetraodon (Tnig), Ciona intestinalis (Cint), fruit fly (Dmel), mosquito (Agam), C. elegans (Cele), Arabidopsis (Atha), and Trypanosoma (Tcru). Vertebrate factors are highlighted in blue, red, and shades of green. (A) Rooted Neighbor-Joining phylogenetic tree generated using ClustalW (1000 bootstraps), based on amino-acid alignment generated by T-Coffee. Bootstrap values are shown. Branch lengths are scaled in arbitrary units. (B) Rooted Gamma-corrected Maximum-Likelihood phylogenetic tree generated using GAMMA and the Phylip program Proml, based on amino-acid alignment generated by T-Coffee. Branch lengths are scaled in arbitrary units.











