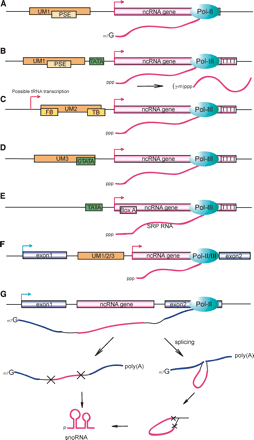
Arrangements of transcriptional elements and genomic locations of small non-coding ncRNA loci, as inferred from genomic and experimental data. (A) TATA-less loci with UM1. This type of locus is characterized by the Upstream Motif 1 and is found both intergenically and intronically. Transcripts from TATA-less UM1 loci generally carry a 5′-end cap, most likely transcribed by RNA polymerase II, and make up biogenesis group I-A, which comprises most spliceosomal snRNAs, a fraction of the SL RNAs, most snlRNAs, and a few C/D snoRNAs along with some unclassified transcripts. (B) Loci with UM1 and a TATA-box. This type of locus combines the UM1 with a TATA-box, and most often a tract of four or more Ts is found within 10 bp of the transcript 3′-terminus. Known RNA polymerase III transcripts like U6 snRNA and RNase P RNA are found at this type of locus. The transcripts may have a single methyl group added at the γ-phosphate post-transcriptionally, as is commonly found in U6 and 7SK snRNAs (Gupta et al. 1990). (C) Loci with UM2. This type of locus comprises a number of both intergenic and intronic snoRNA-like transcripts, along with a few uncharacterized ncRNAs, and makes up biogenesis group II. Transcripts are generally uncapped, and an oligo-T tract is found close to the 3′-terminus, indicating transcription by RNA polymerase III. FB (Front Box) and TB (Tail Box) are the most conserved 15-bp motifs within the 100-bp upstream sequence of these loci, and show strong resemblance to Box A and Box B of the tRNA promoter. A “possible tRNA transcription” initiation site has been indicated to account for the possibility that UM2 is transcribed as a part of the primary transcript (see Supplemental material for details). (D) Loci with UM3. This type of locus has only been found in sbRNAs, and is characterized by UM3, which contains a TATA-box preceded by a strongly conserved G residue. The loci are terminated by an oligo-T tract, and most transcripts are uncapped, suggesting transcription by RNA polymerase III. (E) SRP RNA loci. The C. elegans SRP RNA loci are characterized by a rudimentary TATA-box and a Box A element at ∼10-20 bp downstream of the transcription start, and are terminated by an oligo-T tract. (F) Independently transcribed intronic loci. This type of locus represents subgroups of locus types A-E, in which both the transcribed sequence and the corresponding control elements (promoter, terminator) are found within the intron of a protein-coding gene. This type of locus is found for all the above promoter elements, but is most common for UM1 and UM2 type loci. (G) Motif-less intronic loci. These loci are exclusively made up of snoRNA-like genes, and are often found within an intron of a ribosomal gene. The distance between the ncRNA locus and the preceding exon is generally short (<50 bp) and AT-rich. Transcription is initiated from the host gene promoter, and the snoRNA is processed either directly from the pre-mRNA, or from a spliced intron lariat.











