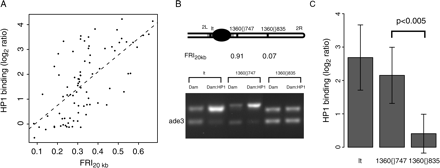
HP1 binding to transposable elements is dependent on flanking repeats. (A) HP1 binding to TEs as a function of the FRI20kb. HP1 binding and FRI20kb for cDNAs on the array that contain sequences homologous to any of the consensus TE sequences (Kaminker et al. 2002) are plotted here. The dotted line shows the linear trendline through the data (Spearman rank correlation ρ = 0.62, P < 2.2 × 10-16). (B) Detection of HP1 binding by DamID and duplex PCR performed on two copies of the same transposable element (1360), both located on chromosome 2R, and the pericentric gene lt, a known target of HP1 (Greil et al. 2003). See Methods for a detailed description of the assay. The top bands correspond to the tested sequence, the bottom bands correspond to ade3, which does not bind HP1 (van Steensel and Henikoff 2000) and serves as an internal standard. (C) Quantitation of the HP1 status. Band intensities were quantified and normalized to the internal standard ade3. Average log2 ratios between Dam:HP1 and Dam reference were calculated for the two transposon copies (n = 5 and n = 6, respectively) and lt (n = 5). Difference between 1360{}747 and 1360{}835 is significant (P < 0.005, Student's t-test).











