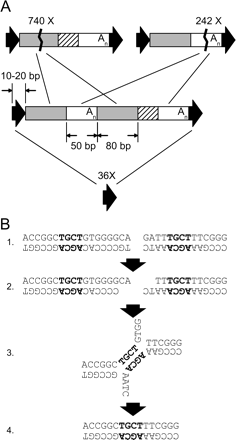
Deletions due to DNA double-strand break repair. (A) Whole and partial Alu element deletions. A full-length Alu is shown in the middle, and black arrows represent target site duplications. Shaded and white internal regions represent internal ∼70% identical homologies. Deletions involving the 84-bp internal Alu homologies (shaded regions) were found 740 times in the human-chimpanzee alignments (top left). Alu internal deletions occurring between the other homologies (white regions) were found 242 times (top right). Precise deletion of entire Alu elements, likely involving the target site duplication (black arrows), was found in 36 cases (bottom) in relatively repeat-free regions since human-chimpanzee divergence. (B) A non-Alu deletion in chimpanzee at human chr1:1448280-1448311. Precise deletions of Alu elements, internal deletions within Alus, and other deletions are explained by an error-prone homology-dependent repair mechanism, involving (1) a double-strand DNA break, (2) resection of DNA and exposure of 3′ tails, (3) homology search, and (4) ligation. In this case, a 4-bp homology mediated a 16-bp deletion.











