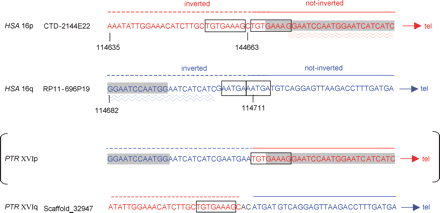
Positions of the breakpoints at the nucleotide level. The sequence in blue represents a portion of HSA BAC RP11-696P19 (16q). The sequence in red is derived from BAC CTD-2144E22 (HSA 16p). The boxes indicate the relative positions of the octanucleotide repeat that flanks the HSA16p breakpoint and the pentanucleotide repeat at 16q. A 22-bp direct repeat found at the breakpoints on chromosomes 16p and 16q is underlined by wavy lines. The HSATII satellite sequences are underlined in gray. The analysis of the chimpanzee sequence in PTR XVIq revealed the presence of two additional nucleotides (AC, in green). Parentheses serve to indicate that the breakpoint region at PTR XVIp, within the HSATII satellite DNA, was inferred from the human sequence, and was not sequenced in PTR owing to the abundance of repetitive satellite sequences.











