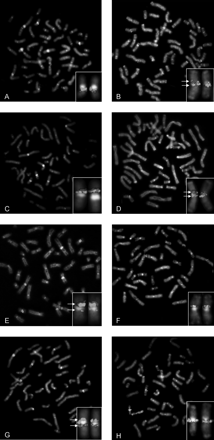
Comparative FISH analysis of breakpoint-spanning BACs (arrows indicate split signals). (A,C,E,G) Human metaphases. (B,D,F,H) Chimpanzee metaphases. (A,B) FISH performed with HSA BACs RP11-696P19 (green) from 16q11.2 and RP11-44I10 (red) from 16q12.1. (C,D) HSA BACs CTD-2144E22 (green) and RP11-598D12 (red), both from HSA16p. In PTR, BAC CTD-2144E22 is split by the inversion. (E,F) PTR BAC RP43-007E19 (green) and HSA BAC RP11-347N4 (red) from HSA 16p12. PTR BAC RP43-007E19 is breakpoint-spanning, since signals are evident at HSA 16p11.2 and 16q11 (E). On PTR XVIp, a single signal of PTR RP43-007E19 is present (F). (G,H) PTR BAC RP43-001I03 (green) and HSA BAC RP11-46D6 (red). PTR BAC RP43-001I03 hybridized to a single location on PTR XVIq (H) but was split by the inversion on HSA 16 (G).











