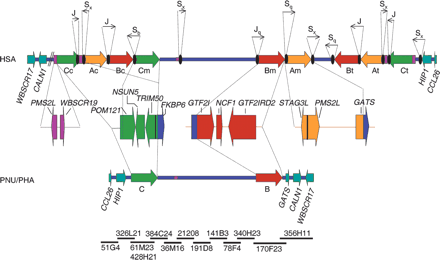
Schematic representation of the genomic structure of the WBS deletion region with the flanking segmental duplications in humans (HSA), and the homologous region in baboon (PNU/PHA). The large blocks of segmental duplications in the human map (A in yellow, B in red, C in green) are represented by thick arrows to indicate their relative orientation with respect to each other. They are exclusively present in the human map, whereas the baboon's genome contains the ancestral loci as single-copy and no large segmental duplication. The blue line represents the single-copy region, and the genes located immediately outside the region in both directions are represented as light blue arrows indicating the transcriptional direction. Some of the multiple-copy modules present in other chromosome 7 locations are shown in purple. The composition of each duplicated block with the corresponding transcriptional units is shown below the human map. Black ovals represent the Alu repeats located at the edges of the segmental duplications in the human map, with arrows indicating their orientation and approximate size (either partialor full Alu elements) shown on top. Note that the entire region including the ancestral loci of the segmental duplications is inverted in baboon with respect to the flanking genes. To define the baboon genomic structure, a clone contig with sequenced BACs from the RP41 library available in public databases has been assembled (NISC Comparative Sequencing Initiative), shown at the bottom.











