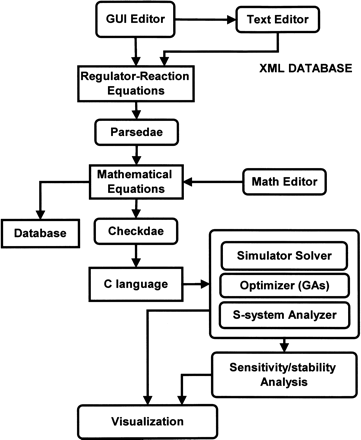
A process flow of the conversion of a biochemical network map to mathematical equations in the CADLIVE Simulator. The parsedae module parses the regulator-reaction equations that are described by the GUI Editor, converting into mathematical equations (checkdae file). The checkdae module converts the mathematical equations into the user functions available for the C language and the parameter file that is used for input of the initial concentrations of the species, the kinetic parameters, and the control parameters for the simulator. These functions and parameters are compiled to link to the simulation engine. In the checkdae file, one is allowed to edit mathematical models directly, because the checkdae file is written in the text format that consists of the variable declaration part and the equation part. The simulator supports dynamic simulation, steady state analysis, S-system analysis, and optimization of kinetic parameters.











