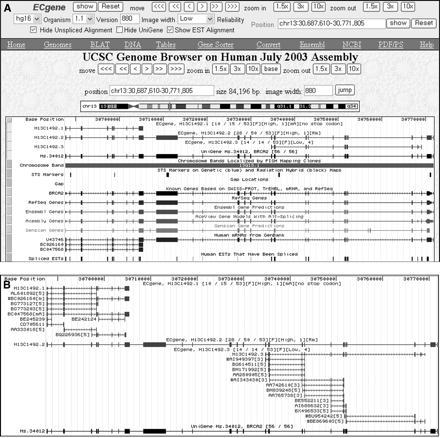
ECgene genome browser. (A) Dense view showing the gene structure. The ECgene ID “H13C1492.1” indicates that the gene is the 1,492nd cluster located on human chromosome 13. The variant number is appended after the ECgene ID. The title line has additional information. “[10/15/53][F][High, 1][mA][no stop codon]” means that this cluster has 53 sequences. The first variant has 15 sequence members, 10 of which are multi-exon clones. The transcript is on the sense (+) strand. It contains mRNA sequence and has polyA evidence. [High, 1] means that the transcript belongs to the ECgene Part A, and the number of MinClones = 1. (B) Expanded view showing sequence alignment. The first variant has a polyA tail on BC047568 mRNA. The third variant belongs to the ECgene Part C (Low reliability) with the number of MinClones = 4. Representative clones belonging to the minimal set are indicated with the “#” sign in front of the accession number. Information on the EST read direction and the presence of mRNA or polyA is appended to the accession number. The browser supports an option of viewing unspliced alignments. If the option of showing EST alignment is unchecked, it will show just the transcript models in a single track. The navigating bars provided in the upper window should be used to make a query to our database. Otherwise, the data in the custom tracks do not change.











