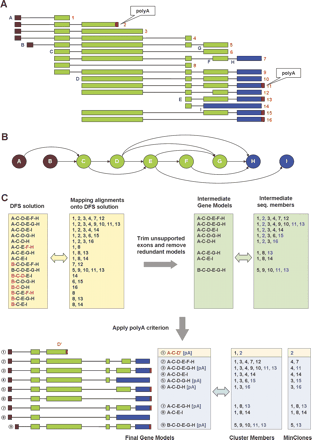
Transcript assembly procedure based on the graph theory. (A) Example of genomic alignment of multi-exon sequences comprising an ECgene cluster. Exons are marked as A, B, C,..., and sequences are numbered as 1, 2, 3,... Exons A and B represent an example of alternative transcription start sites. Exons D, E, F, and G show exon-skipping events, whereas exons F and G occupy the same genomic loci with different 3′ splice sites (acceptor splice site variation). Sequence #14 shows an example of intron retention at exon I. PolyA tails are indicated as small red boxes; they do not align onto the genome. (B) Directed acyclic graph (DAG) representation of genomic alignment. Nodes and edges represent exons and introns, respectively. Exons are colored according to the type of nodes. Source nodes with outgoing arrows only are shown in brown, and terminal nodes with incoming arrows only are shown in blue. Internal nodes are colored green. (C) Transcript models and sequence members. Transcript models in the yellow boxes are the initial solutions from DFS (depth first search) that starts from one of the source nodes and ends with one of the terminal nodes. After mapping sequences onto the DFS solution, unsupported exons (indicated in red) are trimmed off and redundant transcript models are removed. This produces the intermediate gene models shown in green boxes. Then we examine sequences with a polyA tail (shown in blue letters) and ascertain that each transcript has only one polyA site. Truncation at the polyA site in sequence #2 creates a new exon, D′. Final transcript models and sequence members are shown with the MinClones. For example, the third transcript model (A-C-D-E-G-H) is a concatenation of ESTs #4 and #11, and the number of MinClones = 2.











