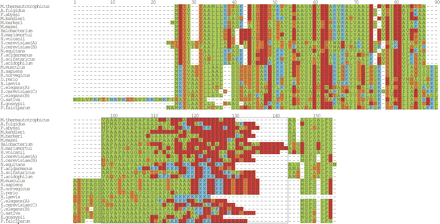
Multiple sequence alignment of the ribosomal protein L12. A multiple sequence alignment of the glutamic acid repeat in L12 from the prokaryote species M. thermautotrophicus str. Delta H, Archaeoglobus fulgidus DSM 4304, Pyrococcus abyssi, M. kandleri AV19, Methanosarcina barkeri, Methanosarcina mazei Goe1, Halobacterium sp. NRC-1, Haloarcula marismortui, Haloferax volcanii, Nanoarchaeum equitans Kin4-M, Ferroplasma acidarmanus, Sulfolobus solfataricus, Thermoplasma acidophilum, and the eukaryote species M. musculus, H. sapiens, R. norvegicus, D. rerio, C. elegans (A) gi 25141400, (B) gi 17543850, S. cerevisiae (A) gi 171813, (B) gi 171815, (C) gi 236358, O. sativa, Eremothecium gossypii, P. falciparum. The boxed region from positions 98–144 highlights the two amino acid-rich regions, the N-terminal alanine-rich region and the C-terminal glutamic acid-rich region. Since no obvious alignment could be built of this region, the sequences were flushed left. The regions to the left and right of the boxed region was aligned with T-COFFEE (Notredame et al. 2000) and then manually adjusted. The final figure was generated with ALSCRIPT (Barton 1993).











