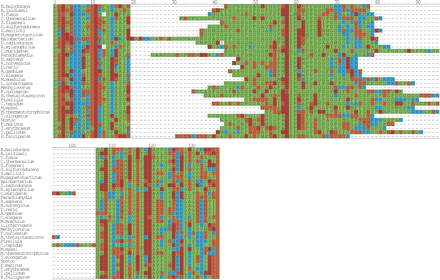
Multiple sequence alignment of DnaJ. A multiple alignment of the glycine repeat in DnaJ (Hsp40). From prokarya: Bacillus halodurans, Clostridium thermocellum, Sinorhizobium meliloti, Shigella flexneri, Magnetospirillum magnetotacticum, Geobacter sulfurreducens PCA, Leptospira interrogans serovar lai str. 56601, Thermosynechococcus elongatus BP-1, Nostoc sp. PCC 7120, Prochlorococcus marinus, Trichodesmium erythraeum IMS101, Methanosarcina mazei Goe1, Treponema pallidum, Rubrobacter xylanophilus DSM 9941, Fusobacterium nucleatum subsp. polymorphum, Methanothermobacter thermautotrophicus str. Delta H, Pirellula sp., Chlamydia muridarum, Parachlamydia sp. UWE25, Bacteroides thetaiotaomicron VPI-5482, Acholeplasma laidlawii, Chlorobium tepidum TLS, Thermobifida fusca, Halobacterium sp. NRC-1, and Deinococcus radiodurans. From eukarya: H. sapiens, R. norvegicus, M. musculus, D. rerio, A. gambiae str. PEST, C. elegans, and P. falciparum. The boxed region from positions 20–106 highlights the glycine/phenylalanine-rich region. The sequences were manually positioned with respect to the first instance of the highly conserved motif DxF (boxed, position 58–60). The regions to the left and right of the boxed region were aligned with T-COFFEE (Notredame et al. 2000) and the final figure was generated with ALSCRIPT (Barton 1993).











