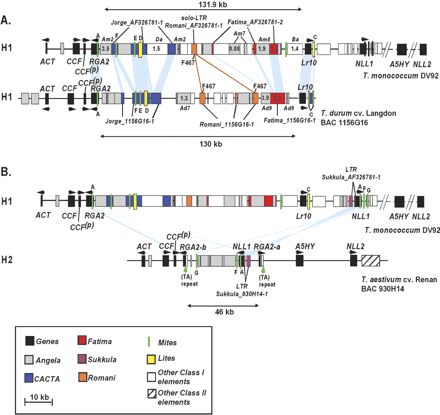
Schematic representation of the sequence organization and detailed comparison at the Lr10 orthologous loci in diploid, tetraploid, and hexaploid wheat. (A.) Organization of the orthologous loci in T. monococcum and T. turgidum and comparison of the genomic region between RGA2 and Lr10 (haplotype H1). (B.) Organization of the orthologous loci in T. monococcum and T. aestivum and comparison between T. monococcum (H1) and T. aestivum (H2) in a region delimited by the two RGA2 fragments of the T. aestivum sequence. Light-blue areas indicate conservation between the orthologous regions in the RGA2–Lr10 interval. The genes are indicated by black boxes. The short arrows located above or below the genes indicate the transcriptional orientation. Different repetitive elements are displayed with different colors. Full names of conserved retroelements are given according to the complete annotation of the BAC sequences AY663391 and AY663392. Am2, Am7, Am8, Ad7, and Ad9 correspond to Angela retroelements Angela_AF326781-2, Angela_AF326781-7, Angela_AF326781-8, Angela_1156G16-7, Angela_1156G16-9, respectively. Da and Ba correspond to Daniela_AF326781-7, Barbara_AF326781-7, respectively. The foldback elements (MITES and LITES) are only given initials in bold type font. (A) Athos; (F) Fortuna; (E) Eos; (D) Deimos; (C) Charon. F467 is a RFLP probe derived from the diploid sequence (Wicker et al. 2001) corresponding to an LTR fragment of a Romani retroelement, and the red lines between the diploid and tetraploid sequences indicate that these LTRs are derived from a common ancestor element. Green arrows indicate the (TA) repeats. Numbers in the boxes indicate the estimated insertion dates of the respective retroelements in million years ago. Numbers below the sequences indicate intergenic distances in kilobases delimited by double-headed arrows. The sequence in the region delimited by dashed lines at the right end of the T. monococcum sequence was only low-pass sequenced, and the intergenic distances in this region were as estimated by Stein et al. (2000). H1 and H2 indicate the two haplotypes, H1 and H2, according to Scherrer et al. (2002).











