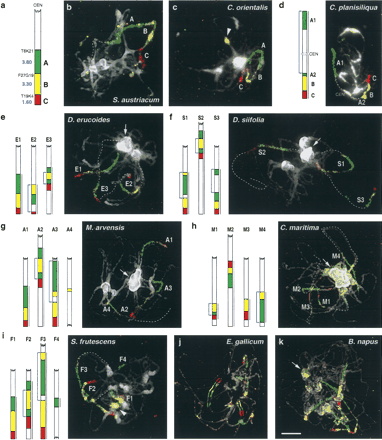
Comparative chromosome painting using the At4-b Arabidopsis BAC contig. (a) Diagram of the At4-b contig from the bottom arm of A. thaliana Chromosome 4 hybridized to pachytene chromosome complements of species shown in b to k. Starting BAC clones and size (megabases) of subcontigs A, B, and C are shown on the left. Subcontigs A, B, and C were visualized as Alexa 488 (green), Cy3 (yellow), and Texas Red (red) fluorescence, respectively. (b,c) One copy of the At4-b contig in Sisymbrium austriacum and Conringia orientalis. The homeologous segments have the Arabidopsis-like structure. (d) One rearranged copy of the At4-b contig in C. planisiliqua. Subcontig A is split into two parts (A1, A2) likely because of a pericentric inversion. (e–i) Three copies of the At4-b contig in (e) Diplotaxis erucoides, (f) D. siifolia, (g) Moricandia arvensis, (h) Cakile maritima, and (i) Sinapidendron frutescens. (j) Six copies of the At4-b contig in Erucastrum gallicum. Whereas four copies apparently show the Arabidopsis-like structure, two homeologous regions bear an inversion (marked by drawing). (k) Six copies of the At4-b contig in Brassica napus; chromosome clustering does not allow unambiguous tracing of individual fluorescence signals. Chromosomes were counterstained with DAPI. Diagrams were drawn to scale (according to measured regions labeled by a bar on the right) for the corresponding species. Blue arrows in diagrams indicate assumed inversions. Strongly DAPI-stained (peri)centromeric heterochromatin (arrows) and nucleolus organizers (arrowheads) show cross-hybridization with sequences of subcontig B (yellow) in some species. Scale, 5 μm.











