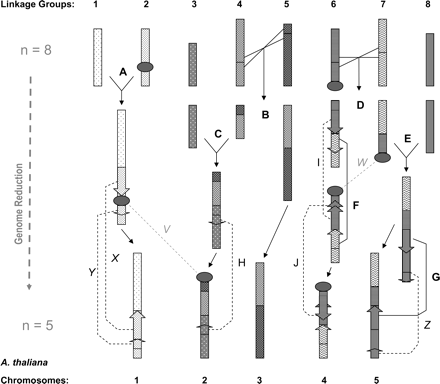
Genome reduction in A. thaliana: Parsimony solution of observable chromosomal rearrangement steps. Linkage groups 1–8 of A. l. lyrata, represented as patterned blocks, are shown across the top of the figure; A. thaliana chromosomes I–V at the bottom. A–G: Seven steps that are found to be in common between A. thaliana–A. l. lyrata and A. thaliana–Capsella spp. genome data; namely B and D, reciprocal translocations; A,C,E, fusion events; F and G, inversions. These seven steps could have occurred in any sequence during genome reduction in A. thaliana's evolutionary history. (H–J) Inversions seen only in the A. thaliana–A. l. lyrata comparative map. (V–Z) Rearrangements seen only in the A. thaliana–Capsella spp. comparative map, namely X,Y, Z, inversions and V and W, translocations of NORs (V and W not included in statistical analysis).











