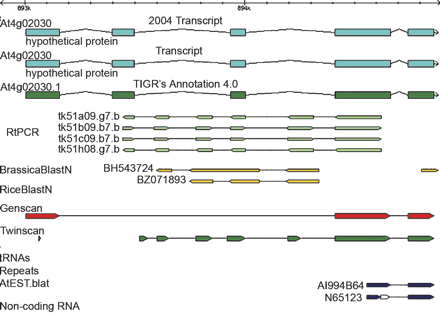
This figure shows a mistake in the Arabidopsis annotation. The image was created by Gbrowse (Stein et al. 2002). The x-axis on the top is the Arabidopsis genome coordinates. The label of each track is listed on the left side, in bold, over the figure. The track “Transcript” corresponds to the March, 2003 annotation. The At4.0 annotation does not contain the correct annotation, possibly because there are no ESTs (track AtEST.blat) in the region where the discrepancy occurs. The figure also demonstrated that the BLASTN alignment of Brassica sequences is often enough to tell whether an exon is present or missing. Twinscan was able to predict some of the missing exons; however, it failed to annotate them all correctly.











