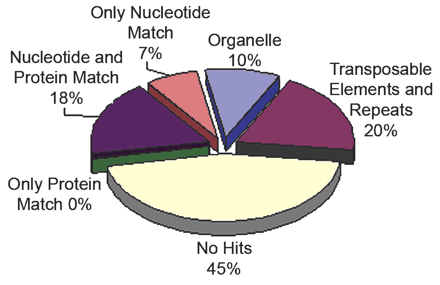
BLASTN, TBLASTX, and BLASTX were used to align 595,321 Brassica oleracea reads against databases of Arabidopsis thaliana chloroplast and mitochondrial DNA, known repeats and transposable elements, and finally, the Arabidopsis thaliana genome and protein sequences. Top BLAST hit was used to classify the Brassica oleracea reads. The reads had to match with an e-value < 1e-10 to be considered as a significant match. The protein database consists of translations of all predicted proteincoding genes in the genome annotation. Therefore, the 7% of the Brassica reads that match only the genome, represent either noncoding RNA or genes that have not yet been annotated.











