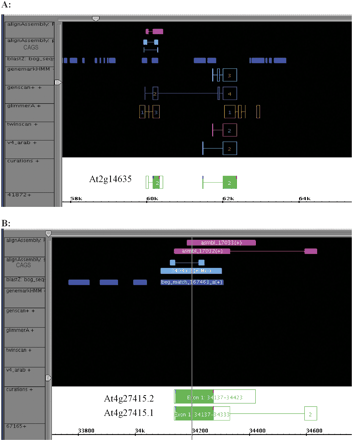
Screen shots of new genes modeled based on RACE results. The images are taken from Annotation Station software. (From top to bottom) The tracks in the gene viewer show the spliced alignment of the assembled sequences from the RACE products (alignAssembly); the location of the paired CAGS and the primers used to test for its expression and subsequently for performing 5′ and 3′ RACE reactions (alignAssembly CAGS); the location of all CAGS in this region (blastZ:bog_seq); predictions of genemarkHMM, genescan+, glimmerA, and Twinscan; version 4 of the TIGR Arabidopsis annotation and the current curations of gene models in this region that incorporate the experimental evidence derived from this study; and the TIGR BAC identifier (the final + sign showing on some of the evidence lines indicates the strand of the alignment). (A) Screen shot of evidence relating to AGI locus At2g14635. The gene model on the left (At2g14635) did not exist prior to this work. Genemark.hmm made no prediction, and other predictions were discordant. The current working model was created based on the alignment of the assembly produced from the sequenced RACE products. (B) Screen shot of evidence relating to AGI locus At4g27415. There are no gene predictions in this region and no working model in version 4 of the TIGR Arabidopsis annotation. The sequenced RACE products generated two assemblies that resulted in the creation of two splice isoforms (At4g27415.1 and At4g27415.2).











