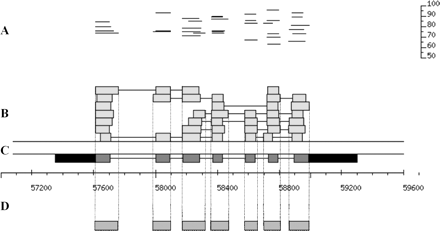
Alignment of Brassica WGS reads with Arabidopsis genomic DNA in the region of At1g20090, a gene whose structure is supported by full-length cDNA evidence. (A) The upper part of the figure shows the length and percentage identity for each BLASTZ alignment segment of Brassica sequence to the Arabidopsis genome after filtering by percentage identity and match length (see Methods). Note that occasionally two overlapping HSPs with the same percentage identity will appear merged into a single line. (B) Each box corresponds to a BLASTZ alignment shown in A. Boxes joined by black lines indicate multiple alignment segments within a single Brassica WGS sequence. (C) The dark and light boxes (expressed sequence) joined by the black line (introns) show the gapped alignment of the experimental cDNA to genomic DNA, with the coding sequence represented in gray and the UTRs in black. (D) The boxes below the BAC nucleotide scale show the extent of the CAGSs constructed from these sets of overlapping BLASTZ alignments.











