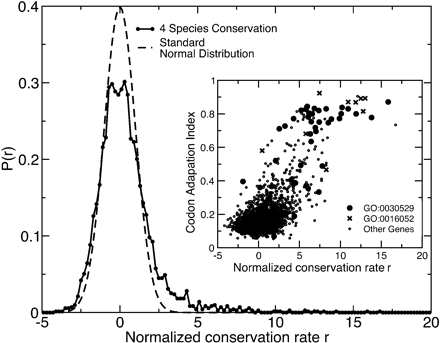
Distribution of normalized conservation rates based on the fourfold sites in each gene. Conservation among all four species S. cerevisiae, S. paradoxus, S. mikatae, and S. bayanus. Each rate has been normalized based on the distribution expected if each fourfold site in the genome were to have an independent and equal probability of conservation at the genome-wide mode rate. The expected distribution if this hypothesis were true is a standard Normal distribution, which is shown for comparison. (Inset) Normalized conservation rate among four yeast species versus codon adaptation index. Most genes with high conservation rates at their fourfold sites have strong codon usage biases. These genes are largely ribosomal (Gene Ontology category GO:0030529, Ribonucleoprotein Complex) or are associated with energy generation (GO:0016052, Carbohydrate Catabolism).











