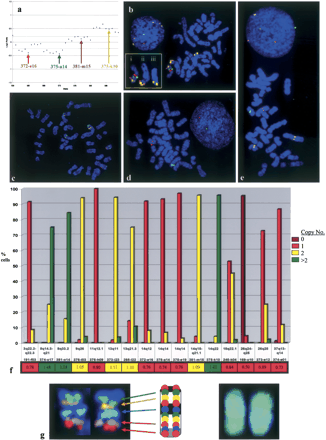
(a) Enlarged partial aCGH profile from Fig. 2B, showing 36 BAC clones distributed along the length of CFA 14. The pattern of fluorescence ratios indicates a copy number loss of the proximal region of CFA 14, interrupted by a small region with a normal copy number. A copy number gain is observed for clones within CFA 14qdist. (b) SLP analysis of four clones from CFA 14 supports these findings. Two proximal clones, 372-e16 (14q12, labeled red) and 375-a14 (14q14, green), are both present in a single copy on one arm of a novel metacentric chromosome in >90% of cells analyzed (shown as “i” in the inset). Two distal clones on CFA 14, 381-m15 (14q15-q21.1, labeled purple) and 375-k10 (14q22, yellow), are also present on the same derivative chromosome arm, suggesting a centric fusion event involving a grossly intact copy of CFA 14. Both distal clones are also present within a second novel chromosome structure (shown as “ii” in the inset). Clone 381-m15 has a normal copy number in >95% of cells, whereas chromosome duplication and translocation have generated a third copy of clone 375-k10 in >96% of cells (shown as “iii” in the inset). Inset: the three derivative chromosomes from this spread that contain segments of CFA 14. (c) SLP analysis of two clones from each of CFA 8 (374-o17, labeled yellow; 381-014, green) and CFA 13 (373-i23, red; 265-l22, blue). This figure shows one of the 15%-25% of cells in this tumor with two copies of both CFA 8 clones. Although all four probes show a normal copy number, there is clear indication of structural rearrangements involving both CFA 8 and CFA 13. (d) SLP analysis of clones from CFA 3 (191-f03, red), CFA 18 (245-k04, green) and CFA 37 (374-e01, purple) demonstrated a single copy number, correlating with observations from aCGH. A fourth probe, clone 169-o10 (CFA 26, labeled yellow) indicated a homozygous deletion (i.e., no visible signal) in > 95% of cells studied, which is also apparent from aCGH analysis. (e) SLP analysis of clones from CFA 11 (376-h09, red), CFA 14 (375-e19, purple), and CFA 26 (373-e12, green) demonstrated a single copy number. Clone 372-i12 (CFA 9q26, yellow) is present as two copies, correlating with the normal aCGH profile; however, it is clear from the position of the probe signals that CFA 9 has undergone structural rearrangement. (f) Combined summary of the aCGH and SLP analyses for these 16 loci. The frequency of probe signals (n = 0, 1, 2, or >2) in the cell population is plotted against the corresponding locus. Beneath each BAC address the aCGH ratio of each locus is indicated and color-coded according to the threshold of copy number: loss (red), normal (yellow), gain (green). In each instance there is agreement between the copy number data derived from both approaches. (g) Application of a chromosome-specific tiling panel in FISH analysis. The central feature shows a CFA 38 ideogram and the cytogenetic location of 10 BAC clones that have been mapped to this chromosome. When all 10 of these clones are labeled with the same fluorochrome, cohybridization results in painting of the chromosome as shown on the right. Conversely, if individual clones are labeled with different fluorochromes, the pool of fluorescently labeled BAC clones (in this instance, five clones) may be used to generate a multicolored chromosome-specific “tiling-set” for the study structural rearrangements (left).











